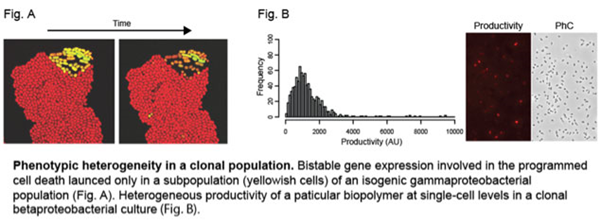
In a classical way, clonal bacterial populations have been thought of as millions of uniform cells, exhibiting little or no phenotypic diversity. However, recent studies using single-cell based approaches revealed such an idea is just an oversimplification. Rather, a pool of isogenic bacterial cells can contain phenotypically distinct subpopulations, which may arise either from stochastic variation in several physiological processes or deterministic reactions such as feedback regulations.
Our goal is to unveil molecular mechanisms underlying phenotypic diversity and reveal physiological functions and evolutionary roles of such heterogeneity in a clonal population. To address the questions, we use molecular genetics and single-cell live imaging techniques combined with high-throughput technologies. Furthermore, we would propose a novel bioengineering scheme based on such cellular potentials at single-cell level.