The main interest of our research is in “phenotypic heterogeneity” in a clonal bacterial population. In classical microbiology, clonal populations of bacterial cells growing in steady environments have been thought of as billions of largely uniform cells, exhibiting little phenotypic diversity. However, recent studies using single-cell based approaches revealed that such an idea is just an oversimplification. Rather, an isogenic bacterial population can contain phenotypically distinct subpopulations, which arise either from stochastic variations at several physiological processes or deterministic reactions such as feedback regulations. Our goal is to unveil molecular mechanisms and evolutionary roles of phenotypic heterogeneity in microbial populations.
Molecular mechanisms of phenotypic heterogeneity and stochastic noise

Individual bacterial cells often exhibit remarkable differences in phenotypes, even though they are genetically identical and proliferate in a steady environment. Why? Because the number of biomolecules within a cell is limited and dynamically changes during a given time scale, biochemical reactions are probabilistic. Such stochastic noise makes all life processes randomly fluctuate. We are interested in the molecular mechanisms which generate, accelerate, and cancel the noise, especially on gene expression. Developing novel systems integrating high-throughput single-cell detection with state-of-the-art technologies for genomics and transcriptomics, we are comprehensively identifying cellular factors and biochemical cascades controlling phenotypic heterogeneity.
Biological roles of phenotypic diversity on horizontal gene transfer
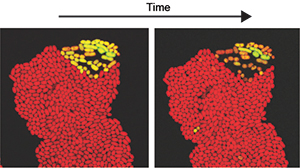
Recent studies on phenotypic heterogeneity have increasingly revealed that phenotypically distinct subpopulations play important roles on microbial adaptation and evolution. In some cases phenotypic diversity could confer robustness on the population to hedge against environmental fluctuation, while in other cases it could promote cooperative functions or division of labor to increase ecological benefits for the microbial species. Working with mobile genetic elements that can horizontally transfer among cells, we recently found a new phenomenon that transfer competent cells arise at a few percent of the population and consequently lyse. This is a good model to study multi-level roles of phenotypic heterogeneity on microbial ecology and evolution, i.e. we ask why only a small subset of the cells become competent? why they need to die? why horizontal gene transfer and mortality are coupling together? there is any trade-off? To address these questions, we are conducting molecular genetics, single-cell time-lapse microscopy, and evolutionary experiments.
Impact of horizontal gene transfer on natural ecosystems

The goal of this project is to unveil evolutionary and ecological roles of horizontal gene transfer (HGT) in natural ecosystems. HGT is a genetic process of microorganisms to transfer genes across phylogenetic lineages. It is believed to be promiscuous in nature and one of the major driving forces for microbial evolution. However, little is known about the frequency of HGT and its contribution to structures, interactions, and functions of natural microbial communities. Using gut microbiota of honey bees as a model ecosystem, we will combine genomics, experimental, and theoretical approaches to investigate dynamics, functions, and consequences of HGT in the bee gut microbiota. The project is an international collaborative research with two other groups and supported by Human Frontier Science Program (HFSP).
Cellular individuality for applied microbiology

In this side project, we are pursuing potentials of cellular individuality for biotechnological and medical applications. A clonal microbial population in principle contains subpopulations which stochastically or accidentally show too high or low activities on any cellular processes, including production of biomass, bioactive substances, or toxic compounds. Controlling such diversity in phenotypic traits, we aim to breed microorganisms for efficient bioproduction and drug discovery.

